Best Practices in Publishing Species Checklists
Version 2.1

Suggested citation
GBIF (2017) Best Practices in Publishing Species Checklists, version 2.1. Copenhagen: GBIF Secretariat. https://ipt.gbif.org/manual/en/ipt/3.0/best-practices-checklists
Cover art credit: Gregory Basco, Black-necked stilt, Himantopus mexicanus
Introduction
This guide provides details on how to utilize the Darwin Core Archive (DwC-A) format as a means to share taxonomic checklist information in a standard way. It focuses on specific components of the Darwin Core Archive format, and some of the supporting extensions to the core taxonomic data class, and provides recommendations on how to best utilize these components to maximize the value of the shared data. This guide does not provide a detailed overview of the Darwin Core Archive format, instead please refer to the Darwin Core Archives How-to Guide.
The DwC-A format and the specific profile described here represent an internationally recognized and ratified data exchange format for sharing taxonomic data. All data exchange standards must strike a balance between the technical scope and capacity on one hand, and social acceptance and uptake on the other. Simple solutions sacrifice coverage and complexity in favour of ease-of-use. Highly complex formats provide more complete solutions for representing any type of data but at the expense of simplicity and require supporting software and expertise. The Darwin Core Archive format represents an intermediate position between the two ends of this spectrum. It focuses on the key elements of taxonomic checklists and enables an enriched set of data types to be linked to this core structure. The data contained in an archive can be readily understood and used by many biologists and data managers familiar with basic structured text files. By providing an international standard that is relatively easy to produce and consume, and that supports many of the key elements that compose a taxonomic data resource, GBIF hopes to provide the creators and managers of checklists with a standardized approach to sharing their data and promote common approaches to the subsequent citation and recognition of their work. A standard format also increases relevance and utility.
Scope
The terms “species checklist” and taxonomic “catalogue” may refer to an overlapping range of taxonomic resources. All of these products contain sets of scientific names that implicitly or explicitly refer to taxa. The set of names included in such a list may be constrained by taxonomic group, geographic region, or by a theme, such as invasive species, or some combination of all three. In order of increasing detail these include the following resource types[1].
-
Name lists – Simple lists of species names with no explicit indication of taxonomic status, but generally implied to consist of accepted names of taxa. Such lists are generally intended to identify a set of taxa included within some regional or thematic context.
-
Nomenclatural lists (Nomenclators) – Lists of names including the nominal taxa, meaning the registry of published usages of scientific names representing nomenclatural acts as governed by the respective Codes of Nomenclature. Most of these acts are ‘original descriptions’ of new scientific names, but other acts may include emendations, lectotypifications, and other acts as governed by the Codes. Synonymy is not included in these lists as taxonomic concept, but only as newly established combination (for botanists) linked to a basionym, thus providing a nomenclatural synonym.
-
Taxonomic checklist – These lists extend nomenclatural lists by adding taxonomic opinion in the form of explicit taxonomic status information and the inclusion of names placed in synonymy. Simple taxonomic lists in this category provide no specific circumscription details regarding the basis for the synonymy. Taxa are often organized into classifications. The term “taxonomic catalogue” may also be used to refer to instances of this, and the remaining categories, particularly if the resource includes verified publication and taxonomic status details.
-
Annotated Checklists - This category extends taxonomic checklists by adding other data types (annotations) to the core, synonymized checklist, such as common names, threat status, distribution and basic descriptive information. When the annotation types provide sufficient detail to effectively define, or circumscribe, a taxon, such as detailed diagnostic descriptions and illustrations, molecular data, specimens, etc., then the annotated list may fall into one of the two categories defined below.
-
Flora or Faunal lists – These are typically books that provide detailed species accounts for a particular region. Details may include many of the data types included in annotated checklists but include specific data types, such as detailed descriptions and illustrations, specimen references and other details that explicitly define (circumscribe) the taxon within the scope of the region which is not necessarily global.
-
Monographs – Monographs are detailed species accounts often published as books for a particular taxon group at the global scale. It will contain detailed nomenclature and synonymy and taxon circumscription details, that include textual descriptions and illustrations, details of specimens examined and included in the definition, and a bibliography of examined literature.
The Darwin Core Archive format with the GBIF Checklist Extensions supports the exchange of key data elements within all of these checklist data types. The specific degree of coverage depends very much on the individual resource. In this document we will use the term ‘checklist’ in the broad sense as a general term for referring to any or all of the resource categories described above. The specific category types will be used when a specific resource is to be referenced.
Darwin Core Archive Format
Darwin Core Archive (DwC-A) is an informatics data standard that makes use of the Darwin Core terms to produce a single, self-contained dataset for checklist data. The collection of files in an archive form a self-contained dataset, which can be provided as a single compressed (Zip or Gzip) file. A dataset is composed of a descriptive metadata document and a set of one or more data files. For more information about DwC-A refer to the Darwin Core Archives How-to Guide.
Checklist Metadata
Documenting the provenance and scope of datasets is required in order to publish checklist data through the GBIF network. Dataset documentation is referred to as ‘resource metadata’ and enables users to evaluate the fitness-for-use of a dataset. It may describe the scope and intended function of the list, methodologies and resources used for its compilation, and the individuals and organizations involved in its creation and management. Metadata is shared in a Darwin Core Archive as an XML document. GBIF provides a metadata profile for species checklists based on the Ecological Metadata Language. A How-to guide describes all the options for describing a species checklist using this format. See GBIF Metadata Profile – How-to Guide.
Checklist Data
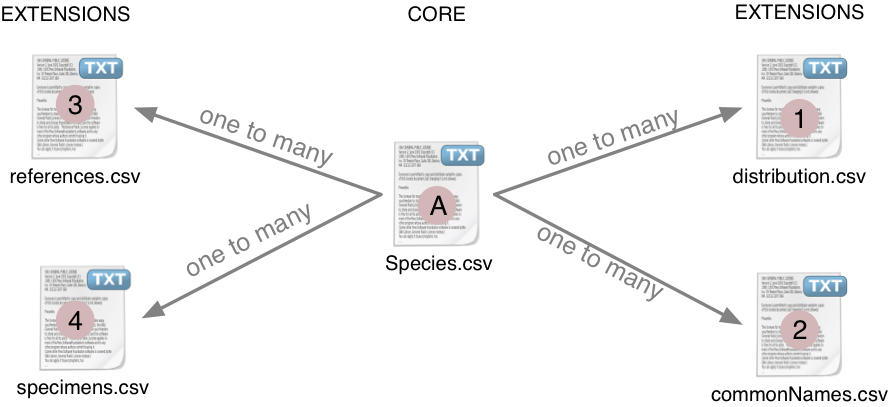
The Darwin Core Archive format provides the structural framework for publishing species checklists. Darwin Core Archives consist of a series of one or more text files, in standard comma- or tab-delimited format. The files are logically arranged in a star-like manner with one core file, containing the basic checklist elements (species list, classification, synonymy) surrounded by a number of ‘extensions’, that describe related data types (such as common names). Links between core and extension records are made using a taxon identifier (taxonID) data element. In this way, many extension records can exist for each single core taxon record. This “star-schema” provides a simple relational data model that supports many types of annotations that are common to species checklists.
Data file formatting recommendations
For ease in understanding, we may use the terms field in this guide to refer to the Darwin Core set of terms in the taxonomic publishing profile to which a users data will be mapped. For example, we will refer to the use of the dwc:scientificName field when referring to the Darwin Core term, scientificName.
-
It is recommended to use tab or Comma-Separated-Values instead of custom field delimiters and quotes.
-
Be careful and consistent with quotation.
-
Encode text files as UTF-8
-
Make sure you replace all line breaks in a data field, i.e.
\r\nor\r\nwith either simple spaces or use 2 characters like$$to replace\rto escape the line break if the intention is to preserve them. Another option is to replace line breaks with the HTML<br>tag. -
Encode nulls as empty strings, i.e. no characters between 2 delimiters, or
\Nor\NULL, but no other text sequence!
Sharing Scientific Names
The Darwin Core supports more than one way to share a scientific name. This includes the following options:
A. Concatenated in the scientificName field
| scientificName |
|---|
Gerardia paupercula var. borealis (Pennell) Deam |
The dwc:scientificName field stores the full scientific name of a taxon including authorship. This field should always be populated with data even if the names are split into component parts (as in C. below). Databases that do not provide a clean separation between the name part and the authorship part of the name should use this field for the entire concatenated name string. This may be needed for hybrid formulas, sensu strictu names, autonyms and other non-trivial binomials. This field is generally used in combination with the dwc:taxonRank field to store the scientific name parts of a full taxonomic list including the higher taxa.
B. Separate Name and Authorship parts
| scientificName | scientificNameAuthorship |
|---|---|
Gerardia paupercula var. borealis |
(Pennell) Deam |
Some databases separate a scientific name into a name part and an authorship part. In this case the dwc:scientificName and dwc:scientificNameAuthorship fields should be used.
C. Separated into name parts
| Genus | specificEpithet | taxonRank | infraspecificEpithet | scientificNameAuthorship |
|---|---|---|---|---|
Gerardia |
paupercula |
var. |
borealis |
(Pennell) Deam |
The Darwin Core provides a series of terms that enable scientific names to be separated into component parts. Some databases store species lists in such parsed components. In this case, sharing data in this form may be an option. If so, however, it is strongly recommended that an additional and complete name be composed from the parts and shared in the dwc:scientificName field (as in section A above). Note that in the table above, the Darwin Core term, dwc:subgenus, is not displayed but represents an additional name component.
Infrageneric Markers
If possible, please provide an infrageneric rank marker as part of the scientific name to avoid confusion with the original / basionym author. For example “Ageratina subgen. Apoda R.M.King & H.Rob” is preferred over “Ageratina (Apoda) R.M.King & H.Rob.” as the later Apoda could interpreted as a subgenus or as the basionym author.
Publishing Classifications
The Darwin Core provides two basic options for publishing classifications or taxonomic hierarchies; normalized and denormalized. These two options account for the primary means by which most classifications are managed in databases.
Normalized Classifications (Parent/Child)
The recommended way to share a classification is in a normalized format. This may also be referred to in a database as a "parent-child relationship" or an "adjacency list". In a normalized taxonomic hierarchy, each taxon is represented by a single row. This includes both species and all higher taxa in the classification. Each row has at least the following component data elements.
-
A dwc:taxonID referring to the current taxon. You can use whatever identifiers you have.
-
The dwc:scientificName of the current taxon. Example: “Panthera tigris”
-
The dwc:taxonRank of the referent taxon. Example: “species”
-
A reference to the taxon identifier of the immediate parent taxon stored in the dwc:parentNameUsageID. In the example below, the parent of record 7, for “Panthera tigris (Linnaeus)” is record 6, the genus “Panthera.”
A sample classification for a single species, the tiger, “Panthera tigris”, is illustrated below. Note that the top-most member of a hierarchy has no parent so that the parent identifier should be empty. Note that dwc:scientificName provides a common field for storing the name in this case but that the full set of options for names is described above in Sharing Scientific Names.
| taxonID | taxonRank | scientificName | parentNameUsageID |
|---|---|---|---|
1 |
Kingdom |
Animalia |
|
2 |
Phylum |
Chordata |
1 |
3 |
Class |
Mammalia |
2 |
4 |
Order |
Carnivora |
3 |
5 |
Family |
Felidae |
4 |
6 |
Genus |
Panthera |
5 |
7 |
Species |
Panthera tigris (Linnaeus) |
6 |
Advantages
-
Efficiency – A normalized classification stores a single reference for each taxon in the hierarchy.
-
Referential integrity – Each taxon component has a distinct identifier that explicitly references its immediate parent. It is easy to verify that the taxonomic hierarchy is complete and properly formed.
-
Extensibility – All taxa are identified with distinct taxon identifiers. This enables higher taxa to be more richly documented through the use of extensions in the same manner as species records.
Disadvantages
-
Convenience - A normalized classification does not provide an intuitive view of the classification hierarchy when viewed in raw tabular form. Many biologists manage classifications in a less efficient, but more visually intuitive, denormalized format, described below. Transforming a denormalized classification to the normalized form is difficult to manually perform.
| A dwc:parentNameUsageID must point to an existing record in the dataset. It is invalid to point to higher taxon identifiers that do not exist as records. |
Denormalized Classifications
This format is familiar to anyone who manages species information in spreadsheets. In a denormalized classification, each row of the data table refers to one of the terminal taxa, such as a species, and a complete set of parent taxa as a set of columns, one for each parent taxon.
This format is not the recommended method for sharing taxonomic data using Darwin Core Archives but is supported by GBIF as it is in common use in many species lists. If this is the method by which data will be shared, it is highly recommended that
-
Each higher taxon column is completely populated. Avoid blanks as in the Plantae example below.
-
Ensure taxonomic integrity of the list. For example ensure that two species in a common genus share the same family. Ensure that if synonyms are included in separate rows, that their classification matches that of the accepted taxon.
| taxonID | kingdom | phylum | class | order | family | scientificName |
|---|---|---|---|---|---|---|
1001 |
Animalia |
Chordata |
Mammalia |
Carnivora |
Felidae |
Panthera tigris |
1002 |
Animalia |
Chordata |
Mammalia |
Carnivora |
Felidae |
Panthera leo |
1003 |
Animalia |
Arthropoda |
Insecta |
Hymenoptera |
Apidae |
Apis mellifera |
1004 |
Plantae |
— |
— |
— |
Poales |
Poa annularis |
Advantages
-
Legibility - The primary advantage of this format is that it is easy to read and the taxonomic hierarchy can be evaluated by simply reading columns.
-
Convenient – Spreadsheet applications and many relational databases make it easy to implement this structure for storing hierarchical data.
Disadvantages
-
Higher likelihood of referential integrity loss – Higher taxa are repeated in this format which can increase the chance that two identical taxa may be spelled differently. Other similar risks are possible with this format. For example it is possible for two instances of the same taxon (example “Felidae”) to be assigned to two different parents, resulting in a conflict of hierarchical integrity.
-
Lack of details for higher taxa – This format treats higher taxa as properties of a species, not as separate taxon records themselves. Therefore, this format does not allow properties of higher taxa to be shared either in the core file or in any extensions.
Other classification-related recommendations
-
Try to include a Kingdom and a nomenclatural code reference for all records even for basic species lists.
-
Try to include Kingdom, Phylum and Family as a minimal classification for denormalized classifications.
-
If it is the same throughout the dataset, consider using a static mapping of the term and value. See the Darwin Core Archive How-to Guide at Darwin Core Archives – How-to Guide for details on mapping global values.
Classification Formats not recommended for publishing
The following examples illustrate data configurations that can fit the profile but are not recommended or supported by GBIF (i.e., GBIF parsers would not handle these cases properly)
-
This example identifies the referent taxon as the last column containing taxon values.
taxonID kingdom phylum class order family scientificName 997
Animalia
998
Animalia
Chordata
999
Animalia
Chordata
Mammalia
1000
Animalia
Chordata
Mammalia
Carnivora
1001
Animalia
Chordata
Mammalia
Carnivora
Felidae
1002
Animalia
Chordata
Mammalia
Carnivora
Felidae
Panthera tigris
1003
Animalia
Chordata
Mammalia
Carnivora
Felidae
Panthera tigris
-
This example attempts is similar to A above but attempts to reduce integrity errors by only recording higher taxon names once
taxonID kingdom phylum class order family scientificName 997
Animalia
998
Chordata
999
Mammalia
1000
Carnivora
1001
Felidae
1002
Panthera tigris
1003
Panthera leo
Please avoid publishing data in these configurations.
Publishing Synonymy
Darwin Core Archive supports the publication of synonyms in species checklists. A synonym is published as a separate record in the core data file. A synonym references the accepted taxon record through the use of the dwc:acceptedNameUsageID field. This field contains the dwc:taxonID representing the accepted taxon record. In the simplified example below, the first record represents the accepted name for a taxon and records 2 and 3 are synonyms.
| taxonID | scientificName | acceptedNameID | taxonomicStatus | nomenclaturalStatus |
|---|---|---|---|---|
1 |
Coeligena helianthea (Lesson 1838) |
1 |
accepted |
|
2 |
Ornismya helianthea Lesson 1838 |
1 |
Homotypic synonym |
|
3 |
Helianthea helianthea (Lesson 1838) J. Gould 1848 |
1 |
Homotypic synonym |
|
4 |
Helianthea typica Bonaparte 1850 |
1 |
Heterotypic synonym |
nomen dubium |
5 |
Helianthea porphyrogaster Mulsant 1876 |
1 |
Heterotypic synonym |
nomen dubium |
6 |
Coeligena helianthea tamai Berlioz & Phelps 1953 |
1 |
Heterotypic synonym |
nomen dubium |
A synonym record is recommended to contain a distinct dwc:taxonID. It must not use the same dwc:taxonID as the accepted taxon record. The simplest representation of synonymy is as provided in the example above where synonyms are listed as distinct records and ‘point’ to the accepted taxon record using the dwc:acceptedNameUsageID. This simple synonymy supports the publication of basic taxonomic checklists with synonym details limited to the core taxon class elements. The dwc:taxonomicStatus field affirms the status of the record. A recommended vocabulary for this field is available. Additional nomenclatural details that may also support the rationale behind the synonymy may be included using the dwc:nomenclaturalStatus field and supporting vocabulary.
Detailed synonymy can be supported by ensuring a unique dwc:taxonID is included in each synonym record and by utilizing the available extensions to support the sharing of checklist annotations. This supports the linking of one or more bibliographic records, specimen records and other data types supported by the GBIF Checklist Extensions to a single synonym record in the core data file. If a dwc:taxonID is not provided for a synonym record, extensions cannot be used as they rely on the dwc:taxonID to provide the link to the taxon record in the core file. A simplified example below illustrates the use of two files (expressed as tables) to provide a bibliography for a synonym using the References extension. The shared dwc:taxonID is highlighted in the example.
Taxon.txt data file
| taxonID | scientificName | acceptedNameUsageID | taxonomicStatus |
|---|---|---|---|
1 |
Coeligena helianthea |
1 |
accepted |
2 |
Ornismya helianthea |
1 |
synonym |
3 |
Helianthea helianthea |
1 |
synonym |
References.txt data file
| taxonID | Bibliographic citation |
|---|---|
2 |
Schmidt, O. 1870. Grundzüge einer Spongien-Fauna des atlantischen Gebietes. (Wilhelm Engelmann: Leipzig): iii-iv, 1-88, pls I-VI. |
2 |
Laubenfels, M.W. De 1942. Porifera from Greenland and Baffinland collected by Capt. Robert A. Bartlett. Journal of the Washington Academy of Sciences 32(9): 263-269. |
Other Synonymy Do’s and Don’ts
-
An dwc:acceptedNameUsageID must point to an existing record in the dataset. It is invalid to point to accepted taxa that do not exist as records.
-
Do not confuse the dwc:higherTaxonID used to describe a classification with the dwc:acceptedNameUsageID used to describe the taxonomic status of a record.
-
Do not “chain” synonyms. A synonym should only point to accepted taxon records via dwc:acceptedNameUsageID. They should never point to another synonym.
Nomenclatural Synonymy
Nomenclatural synonymy is supported in the core data file through the use of the dwc:originalNameUsageID field. This field refers to the row representing the original taxon reference for the name. This record is recommended to provide a bibliographic citation in the dwc:namePublishedIn field, which refers to the publication in which the name was originally established.
| taxonID | scientificName | originalNameID | namePublishedIn |
|---|---|---|---|
1 |
Tetrao afer Müller 1778 |
1 |
J. Syst. Nat 7:31 |
2 |
Pternistes afer (Müller 1778) |
1 |
|
3 |
Francolinus afer afer (Müller 1778) |
1 |
Nomenclatural and taxonomic synonyms may be designated in the same taxon record.
| An dwc:originalNameUsageID must point to an existing record in the dataset. It is invalid to point to accepted taxa that do not exist as records. |
Pro-parte Synonymy
Sometimes the same name may be a synonym for more than one accepted taxon or may be both an accepted taxon name and a synonym. These are caused by splits and circumscription changes where, for example, a series of types may be divided among multiple taxa. The recommended practice for sharing pro-parte synonyms is represented in the example. In this example, Vireo solitarius is an accepted taxon name and it is also included in the synonymy for both Vireo cassinii and Vireo plumbeus. In the case of the synonyms, they are represented as a single record with accepted taxon reference concatenated in the dwc:acceptedNameUsageID field and separated by a pipe (“|”) character.
| taxonID | scientificName | acceptedNameUsageID | taxonomicStatus |
|---|---|---|---|
1 |
Vireo solitarius |
1 |
accepted |
2 |
Vireo cassinii |
2 |
accepted |
3 |
Vireo plumbeus |
3 |
accepted |
4 |
Vireo solitarius |
2|3 |
pro-parte |
| IPT users should define the multi-value delimiter for each source file in the IPT. Refer to Source Data section of the IPT User Manual for additional guidance. |
Citation and Attribution
Taxonomic checklists often represent significant intellectual and financial efforts on the part of the individuals and organizations who compile them. Some checklists may be derived from, or may reference, other source checklists to create new distinct thematic, regional or taxonomic views of the same source authority. Proper attribution and visibility of these sources is therefore a high priority.
The DwC-A format provides a range of options and recommendations for providing proper citation and attribution. This range extends from global citation and attribution information that form part of the resource metadata down to record-level data elements. These options support the provision of multiple levels of attribution.
Metadata Citation and Attribution
The GBIF Metadata profile supports resource-level data elements that contribute to citation and attribution and enable detailed description of the scope and provenance of a checklist. A complete reference list to all the metadata elements is beyond the scope of this document and available but specific citation and attribution-related elements include:
-
Intellectual Property Rights – The metadata profile contains a rights management statement for the resource, or a reference to a service providing such information, such as a Creative Commons license. It also includes an element describing the intended use and purpose of the dataset.
-
Individuals and Organizations – The metadata profile enables the description of any and all individuals, institutions or organizations that may be associated with a dataset. These agents may be ascribed different roles relative to the dataset and may include URLs to each resource. This section provides one method for describing and linking to individuals and organizations that have contributed to a checklist.
-
Source URL – Links to the homepage of the source
-
Project Information – If the checklist is linked to a particular project (e.g., “The Catalogue of Life”) there are a set of fields for describing the project in detail.
-
Citation – This element allows the checklist publisher to specify exactly how the checklist data should be cited when used. Example “Appeltans W, Bouchet P, Boxshall GA, Fauchald K, Gordon DP, Hoeksema BW, Poore GCB, van Soest RWM, Stöhr S, Walter TC, Costello MJ. (eds) (2011). World Register of Marine Species. Accessed at http://www.marinespecies.org on 2011-02-22.”
-
Bibliography – A complete bibliography of sources can be described and included in the metadata document.
Data-level Citation and Attribution
Attribution and citation information recorded in the metadata document is common to all data records in a dataset. In some cases, additional granularity is needed even down to individual records. In these cases, there are record-level terms that are recommended for use in specifying citation and attribution information.
-
dwc:nameAccordingTo : This term can be used to identify the individual or citation that serves as the authoritative taxonomic reference for the record. (Example “Erpenbeck, D.; Van Soest, R.W.M. 2002. Family Halichondriidae Gray, 1867. Pp. 787-816. In Hooper, J. N. A. & Van Soest, R. W. M. (ed.) Systema Porifera. A guide to the classification of sponges.”)
-
dwc:nameAccordingToID: A unique identifier that returns the nameAccordingTo reference as described above. This could be a URL for example.
-
dwc:datasetName: If the record is derived from an external dataset this dataset can be cited as a text string. (Example, “World Register of Marine Species, cited on 12 April 2011”)
-
dwc:datasetID – An identifier that refers to a dataset, preferably resolvable.
-
dc:source – Link to the source web page
Use Case 1 - Checklists composed of multiple contributing datasets (e.g., Catalogue of Life, PESI, WoRMS)
A taxonomic dataset may be a composite of multiple contributing sources, each of which needs to be acknowledged in addition to the collective resource itself. There are many examples of this. Perhaps the largest such collective effort is the Catalogue of Life Annual Checklist which aims to provide a complete listing of all the worlds living species. The checklist itself is composed of individual datasets that represent major taxonomic groups. Each of these resources, in turn, may be composed of contributions from a sub-network of specialists.
Other examples include the Pan-European Species list, which is composed of a number of contributing datasets that include Fauna Europaea, the European Register of Marine Species, Euro+Med PlantBase and others. The World Register of Marine Species represents another such network.
The recommended practice for effectively documenting the provenance of these sorts of resources can be summarized as follows.
-
A single metadata document is created to represent the collective resource itself, (e.g., the Catalogue of Life, the The World Register of Marine Species, etc.) This metadata document provides the proper citation, agents, rights, and other elements identified above. This document filename is referenced the Darwin Core Archive descriptor file, meta.xml. This links the document to the entire DwC-A dataset. Recommended best practice is that this file uses the GBIF metadata profile and be named EML.xml. In this case, the metadata descriptor XML would look like this:
<archive xmlns="http://rs.tdwg.org/dwc/text/" metadata="eml.xml">
-
Additional metadata documents can be created for each of the component datasets and included in the archive. This allows each sub-component dataset to be documented as completely as the “parent” dataset with its own recommended citation, contributing individuals etc. As these datasets do not document the entire collection, they are not referenced in the meta.xml descriptor file. Instead they are referenced from individual data records via the dwc:datasetID term. If the metadata documents are included in the archive itself, the dwc:datasetID equals the filename of the document. Alternatively, it could refer to a URL or some other unique and resolvable identifier for the information. A less recommended but alternative approach would be adding a URL to a simple web page that describes the dataset as opposed to a structured metadata document.
-
To cite individuals at the record level, providing a 3rd level of citation, it is recommended to use the dwc:nameAccordingTo field. Additional record-level terms are provided above.
Use Case 2 - Checklists derived from one or more authority sources
A species checklist in this use case is compiled for a specific purpose but derives it’s basic taxonomic structure from one or more external taxonomic checklists that serve as authority files. The new compilation may include additional annotations to the basic source record that apply to the new lists focus. An example might be a European national species checklist derived from a database such as Fauna Europaea or the Catalogue of Life, which, in principle, provide the complete listing for a country as a subset of their own coverage. A national list may then add additional regional details such as a national threat status or some other property of interest, resulting in a new, derived dataset. In this case, it is important to be able to provide record-level attribution and linkages to the source dataset. The recommended means to do this are as follows.
-
A single metadata document is created to represent the new, derived resource itself (e.g., National Checklist of the Netherlands). Datasets that are referenced can be cited in this metadata document.
-
Fully described as organizations with a role of Contributor and links to the source web site.
-
Cited in the bibliographic section with the citation represented as recommended by the referenced dataset.
-
-
In the data files, additional attribution and linkages can be made at the record-level. This includes:
-
Referencing the dataset by name in dwc:datasetName
-
Referencing the dataset by ID (such as URL) in dwc:datasetID and linking to the dataset home page
-
Providing a link to a corresponding species page on the referenced dataset web site using dc:source
-
If dc:source is reserved for pointing to URL for the derived database, a link to the source database can still be added using the Alternative Identifiers extension.
-
-
If the source dataset provides globally unique identifiers for the taxa referenced in the list, they can be used as the taxonID in the derived dataset. This ensures an explicit link to the source taxon and is highly recommended if available.
-
Use the dwc:nameAccordingTo or dwc:nameAccordingToID to refer to the taxon definition in the corresponding source record as a citation or a URL.
-
Sharing Vernacular Names
The sharing of vernacular name data associated with taxa in taxonomic checklists is supported. Vernacular names are shared as a separate, related file using the Vernacular Names extension. The extension supports a rich set of properties for describing vernacular name usages that include regional and morphological qualifiers.
Vernacular names are referenced via an extension, therefore they must be linked to a named taxon in the parent core data file. It is further recommended that a vernacular names record provide a language reference that identifies the language represented by the vernacular name use. The best practice is to use the ISO 693 language code for sharing language information. Vernacular names may also have distinct regional uses and this can be specified through a dwc:locality element or, at a less precise level, using a dwc:country term. It is recommended that country names utilize the ISO 6133 country codes.
Sharing Species Descriptions
The sharing of descriptive information related to a taxon is supported. Descriptive data are shared as a separate, related file using the via the Taxon Description extension. Descriptive data can be assigned to distinct description types and, as the data is published in an extension, multiple descriptive records may be linked to a single taxon, supporting a relatively rich set of data per taxon. It is recommended that the description type vocabulary be used to describe the descriptive information.
Multi-line descriptions
Descriptive information should be limited to single paragraph text blocks. Multiple paragraphs containing line breaks should be avoided or carefully managed in order to maintain the integrity of the resultant text file output as the Darwin Core Archive. Multi-line data fields served as text files require the record delimiters, which are usually line break characters, to be distinct from the line breaks used within a multi-line field. The best method for supporting multiple lines in a single field is to replace breaking characters with a non-breaking character or character set that a user can replace with proper breaks when the data is parsed and used. One option is to use the HTML break tag <br>.
Sharing Species Distributions
The sharing of distribution data is supported. Distribution data are shared as a separate, related file using the Species Distribution extension. This enables multiple distribution records to be published per taxon. The distribution extension is not only used to designate national or regional distribution descriptions, it also supports the qualification of the referenced distribution in regard to the threat status of the taxon, whether it is introduced, native, etc., and other properties that might be tied to a specific defined area.
The recommended best practice for specifying a distinct area is via a resolve-able or well-known area identifier published via the dwc:localityID element.
If the dwc:country element is used, it is recommended that the ISO 6133 country codes be used.
Sharing References
The sharing of bibliographic citations is supported. Bibliographic data are shared as a separate, related file using the References extension. The References extension is recommended and designed for use in the sharing of synonymy information in monographs and annotated checklists. It supports the sharing of a parsed citation and therefore provides a more granular citation format that some of the citation-storing data elements in the core data file, such as dwc:namePublishedIn. This extension supports the taxonomic and nomenclatural qualification of a reference via the dc:type property, which, when used with the Reference Type vocabulary, can be used to distinguish a set of references related to a taxon.
Note that extensions must link to the taxa in the core data file by providing the taxonID in the taxonID field. See the structure of Darwin Core Archives above.
Sharing Type information
The sharing of information about types and specimens is supported. These data are shared as a separate, related file using the Types and Specimens extension. It supports the sharing of basic information about type specimens, type species and genera.
Sharing Links and Identifiers
It is possible to share and describe multiple links to related external resources using the Alternate Identifier Extension. It allows data publishers to embed links back to the source database or document via resolveable identifiers. Multiple identifiers, perhaps linking to both a web page as well as a more machine-readable web service response, may be provided for a single taxon. It is recommended that a format be included for each record to enable a user to know how to interpret the response information if an identifier is resolve-able. This is usually done by including the media type (or MIME type) in this field. A complete list of media types is available here.
Creating a dynamic link to a species page
Often, a link back to a source database follows a common format, differing only in the identifier number or taxon name used in the URL. This can result in a verbose and bloated extension file. The Darwin Core Archive format supports a more efficient way to define a URL template, which only needs to be defined once, and allows a variable to be embedded in the template eliminating the need for repetitively repeating a set of URLs for each taxon in the data file. This is done via the XML metafile component of a Darwin Core Archive. It does not use the References extension. This requires editing the XML metafile which requires some degree of familiarity with XML. GBIF provides a complete guide the to Darwin Core metafile.
The metafile supports the creation of variables in the metafile that may refer to a web page or web service call. This variable may be embedded in the URL and include a taxon identifier or the taxon name as one of the parameters in the URL. Any column in the published data can be referenced by enclosing the index number in curly braces “{}”. The taxon identifier in the core data file can also be referenced via the variable “{id}.” The following examples illustrate these features:
-
The Integrated Taxonomic Information System (ITIS) uses Taxonomic Serial Numbers (TSN) to provide links to taxon pages on its web site.
If a core data file is published using the ITIS TSN system a link can be composed and tied to the “identifier” term in the core data standard using the following syntax.
<field default="http://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value={id}" term="http://purl.org/dc/terms/identifier"/>where the original numeric value is replaced by the variable
{id}. This value would be derived from the core ID. -
The 2010 Catalogue of Life Annual Checklist provides similar identifiers. It also supports name-based searches that can also be encoded as URLs. For example,
http://www.catalogueoflife.org/annual-checklist/2010/search/all/key/Struthio+camelus/match/1
embeds a scientific name “Struthio camelus” into a URL. Full scientific name combinations can be published in the core data file using the Darwin Core term “scientificName.” If we assume that this term represented the 12th column in our core data file we could use the syntax
<field default="http://www.catalogueoflife.org/annual-checklist/2010/search/all/key/{12}/match/1" term="http://purl.org/dc/terms/identifier"/>where
{12}represents the 12th column value that will be substituted in the URL.
GBIF Checklist Extensions
The core data file in a checklist contains taxon records. The set of terms that can be used to describe a taxon record is defined by the Taxon (Core) Extension.
Each taxon record can be extended with one or more records in an extension file. The set of terms that can be used to describe each extension record is defined by its Extension.
Below is the complete list of Extensions that can be used to provide additional information about a taxon record:
Taxon (Core) Extension
Latest version issued: 2024-02-19
Use this set of terms to provide the fundamental information for a species checklist including classification, synonymy and other key elements. Each row in this list represents a taxon name, either an accepted name or a synonym. The terms in this class support different methods for representing classification information. Classifications can be shared “spreadsheet-style” with columns for Kingdom, Phylum, Class, etc. or they can be shared “database-style” with each taxon row possessing a field containing the ID of its immediate parent. Please note that the tables contain the complete list of acceptable terms. The minimum requirement for sharing a checklist is as little as a list of species although an accompanying ID is highly recommended. Use this list of terms to identify the terms that best match the data to be shared. Don’t be daunted by the term names. Read the description to locate relevant terms.
Vernacular Names Extension
Latest version issued: 2015-02-13
This extension provides the means to share information related to common (vernacular) names linked to taxa in the core data file. Multiple vernacular names can be linked to the same taxon via the taxonID.
References Extension
Latest version issued: 2015-02-13
Use this extension to describe one or more bibliographic references related to a taxon in the core data file. Use the type field to qualify the references. This extension supports the sharing of referenced synonymic checklists.
Species Distribution Extension
Latest version issued: 2022-02-02
Use this extension to share information on one or more distribution references for a taxon. One or more locality records may be linked to the same taxon. For example multiple localities, regions, or countries may be listed. Use this extension to describe the threat status for a taxon, seasonal distribution changes, and other properties linked to a taxon in a particular region.
Species Description Extension
Latest version issued: 2015-02-13
Use this extension to provide descriptive text for a taxon. This is typically in the form of a single paragraph per record as would be normally stored in a database. Descriptions can be qualified by a type to indicate, for example that the description is related to, for example, the morphology conservation, reproduction, etc. Multiple descriptions equal multiple records in a descriptions file.
Alternative Identifiers
Latest version issued: 2015-02-13
Use this extension if you have more than one identifier or link to information about the taxon. A source database, may, for example provide access to the source data records through a web page, a web service, and a resolvable identifier such as LSID, DOI or other means.
Types and Specimens Extension
Latest version issued: 2015-02-13
Use this extension to share data relating to one or more specimens or type references linked to the core taxon
Resource Relationship Extension
Latest version issued: 2022-02-02
This extension is used to describe one or more relationships that the core taxon has with other taxa, either in the source list or included in the record. This extension could be used, for example, to provide a list of plant species (one record per species) pollinated by a bee species listed in the core species list.